
A uniquely advanced solution to prep for mass spec analysis, the platform’s capabilities and streamlined workflows offers advantages over traditional methods like laser capture microdissection (LCM), antibody-based imaging workflows, and proximity labeling.

Extract all proteomic information without antibodies or targeted panels across an entire sample.
350 nm precision at sub-cellular scale.
Use with FFPE/fresh frozen tissue samples or fixed cells.
Discover novel biomarkers or therapeutic targets of disease-associated locations.
Discover low copy number proteins from increased dynamic range due to subcellular protein isolation.
novel proteomes of organelles, cells and organisms.
Two photon guided photo-biotinylation identifies only proteins in regions of interest.
Traditional proteomic technology often relies on predefined panels or antibody-based enrichment, which introduces bias—only known or expected proteins are detected.
But Syncell’s Microscoop® labels and analyzes all proteins in a specific region of interest without pre-selection, allowing mass spec to capture low-abundance or rare proteoforms, and reveal otherwise unknown protein constituents with higher dynamic range and specificity than laser microdissection or proximity labeling methods.
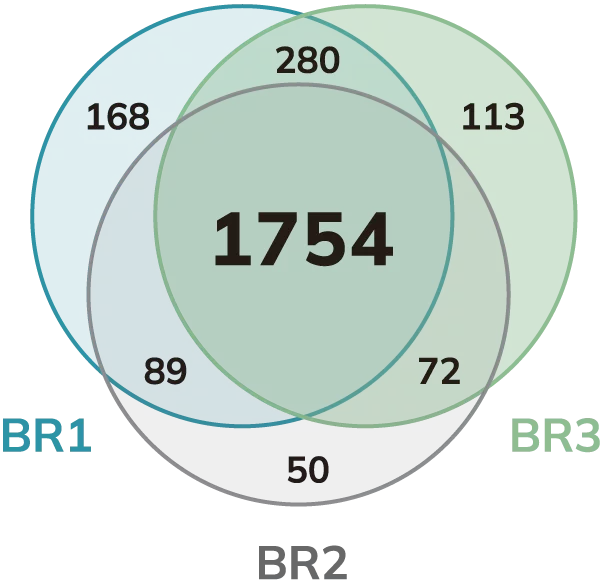
Venn diagram of the stress granule proteins spatially isolated by Microscoop® and analyzed by mass spectrometry.
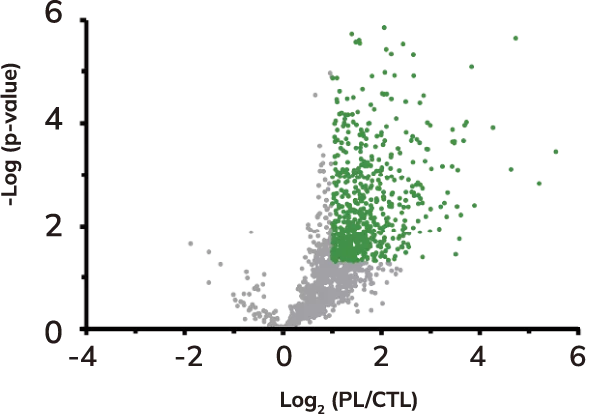
Volcano plot of protein levels in relative photolabeled (PL) samples to control (CTL) samples in log2 scale. Over-represented (enriched) proteins are shown in green.
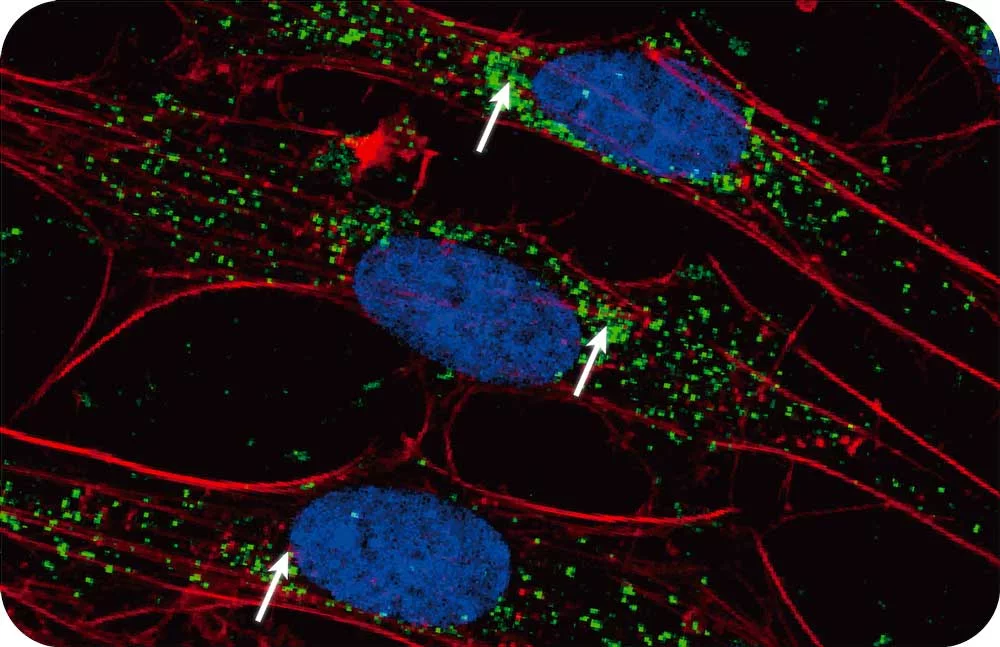
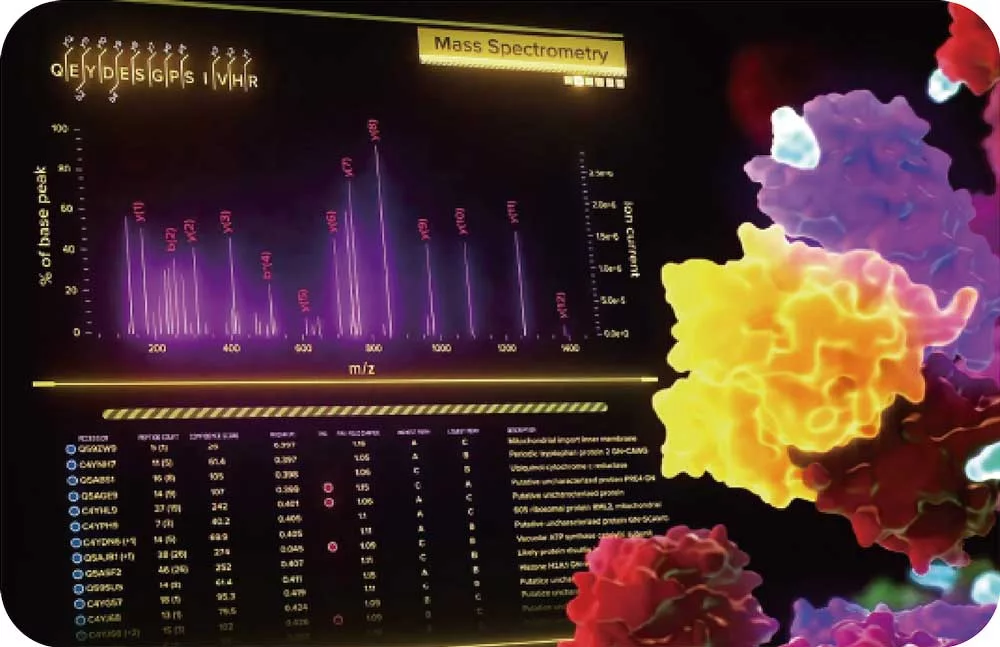
Mass spec alone doesn’t provide spatial information but combining it with the power of Syncell’s microscopy-guided photo-biotinylation technology enables researchers to link spatial location with the entire local proteome allowing you to:
By restricting the proteomic analysis to defined cellular or subcellular regions, our proprietary opto-proteomic platform improves:

Microscoop can work with FFPE/fresh frozen tissue, preserving the integrity of archived clinical samples. This brings MS-based insights into areas where methods like proximity labeling are not capable:
“The enrichment in the data is the best I’ve ever seen.”
-Michael Ford, PhD, Founder, MS Bioworks


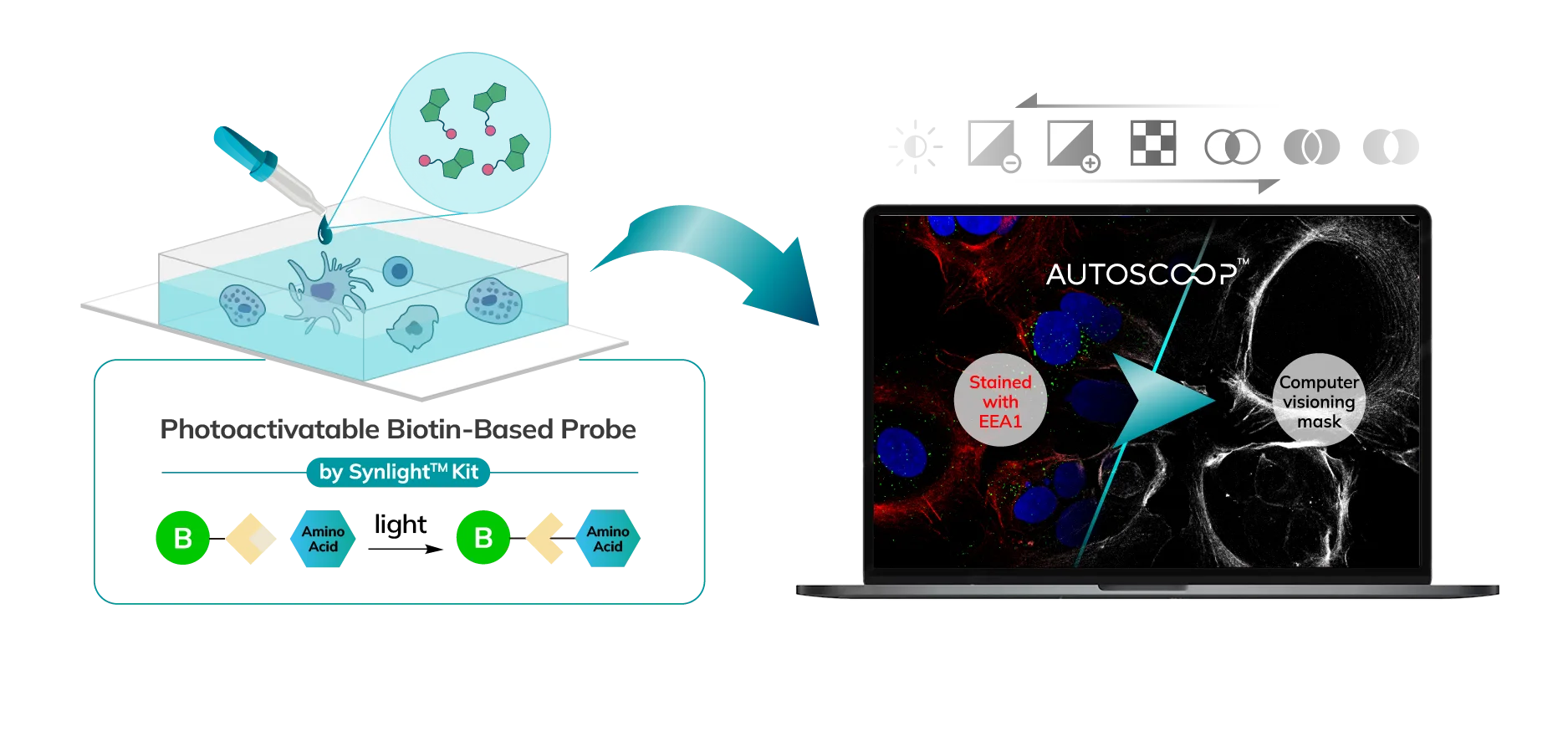
Cells (Fixed, Frozen) or Tissue (FFPE, Fresh Frozen, PFA/Methanol fixed) are stained with a marker that the researcher uses to predefine areas for deeper proteomic analysis. This can be stained with an immunofluorescent marker, multiple markers or other anatomical marker of interest that can be identified via microscopy. One can define these regions based on size, shape, location, distance, and intensity. The user places the stained sample on the Microscoop stage and the sample is imaged. These ROI’s are identified manually within one field of view (FOV) and converted by the Microscoop Autoscoop Software to an image mask, a binary parameter, that tells the system which specific locations to target for whole proteome analysis. The image mask is automatically applied to the entire sample. The ROI’s are the “blueprint” by which the 2-photon laser uses in the next step to target the specific locations of interest.
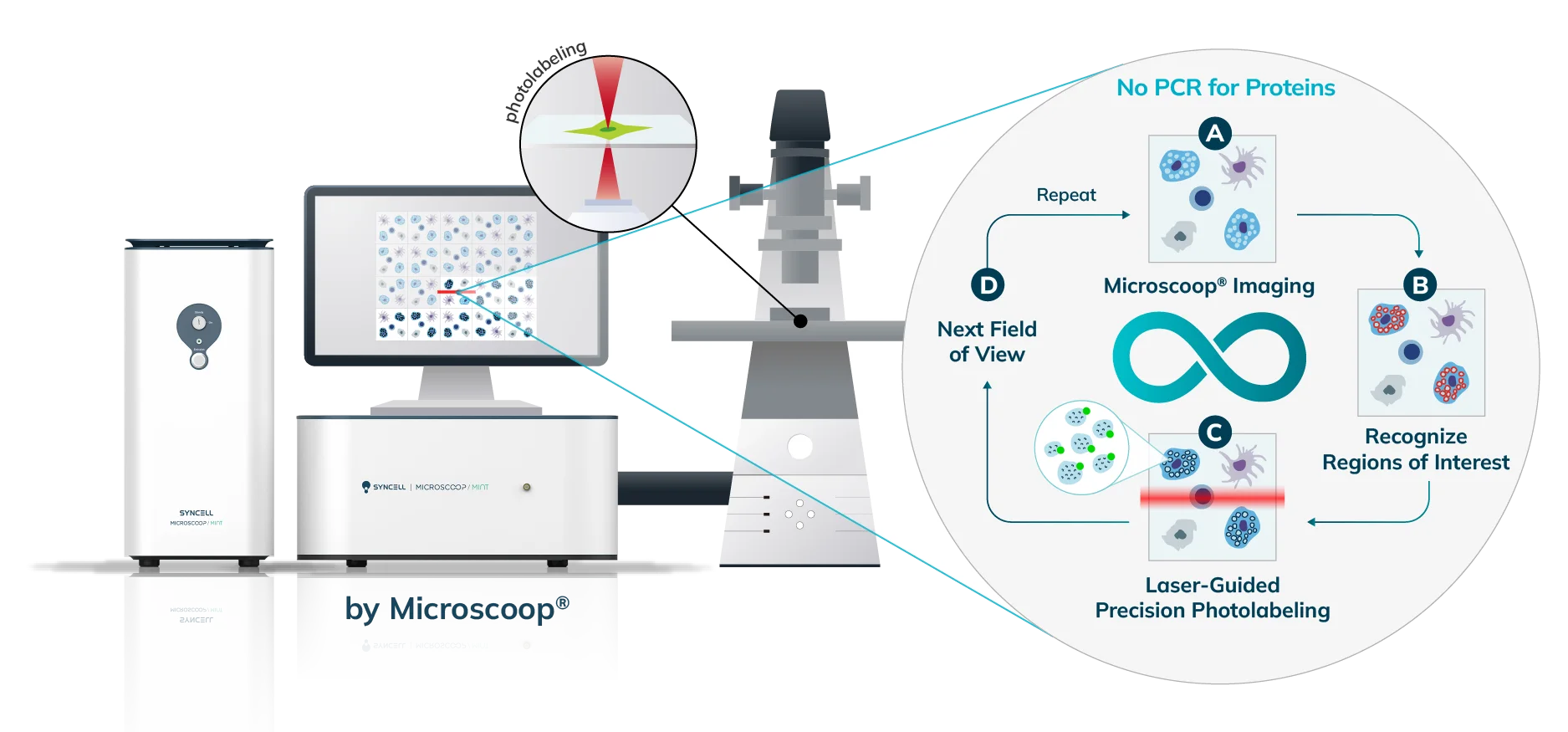
The Synlight Rich kit, containing a photobiotinylation reagent, is applied to the entire sample. A femtosecond light source is controlled mechatronically is used to illuminate the region of interest one pixel at a time. This patterned illumination triggers targeted protein photo-biotinlyation with high spatial precision through the reactions of light-sensitive probes of the Synlight-Rich™ Kit. This patterned photolabeling is repeated for hundreds to thousands of FOVs automatically. The number of photolabeling events is dictated by the size and quantity of the ROI’s. The intention is to aggregate all ROI protein material together to ensure there is sufficient protein to go through the protein extraction and input to downstream mass spectrometry or alternative downstream proteomic readouts. This entire step is done automatically and requires no user intervention until all pixels are labeled by the laser across the FOV’s of interest.
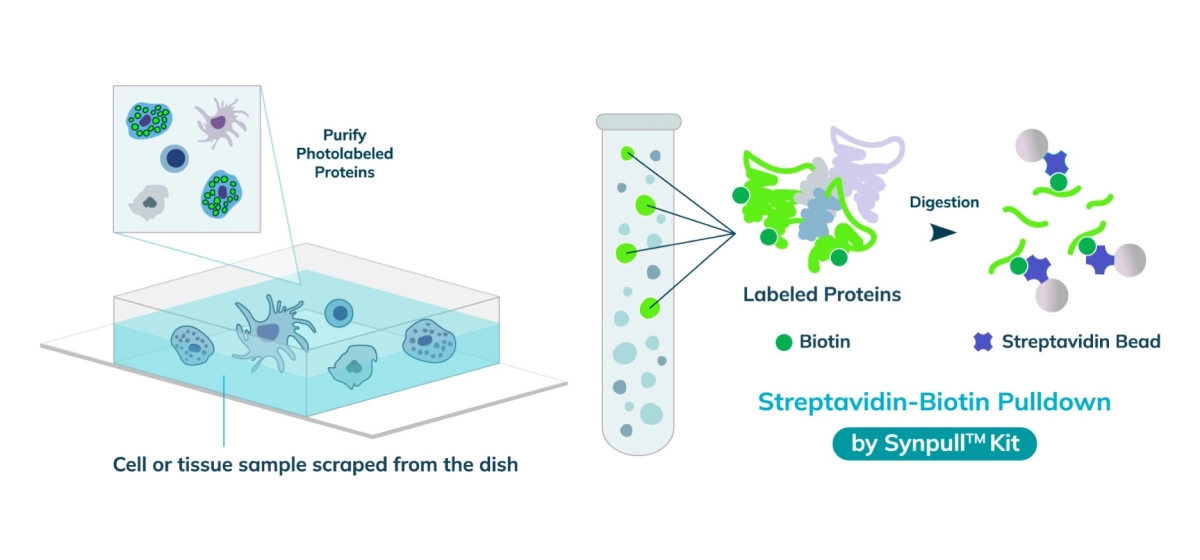
The entire sample contents are collected from the slide or chamber. Materials from multiple slides or chambers can be pooled together to increase the total protein content. The Synpull Kit is used to carry out the next steps of extraction. Using the kit, the samples are lysed, biotinylated proteins are enriched and purified by immunoprecipitation, and proteins digested into peptides for proteomic analysis.

The collected peptides can be lyophilized for downstream proteomic analysis. Typically, an unbiased proteomic analysis of the protein lysate collected from the Synpull Assay is performed on a mass spectrometer (LC-MS/MS). Proteomes of both the photo-labeled and unlabeled (control) samples are obtained. By comparing the proteomic makeup of the control and photolabeled samples, a location-specific proteome is obtained with high sensitivity and, high specificity with high spatial precision. Validation can be done by colocalization of immunostaining, subsequent follow-on mass spec experiments or additional functional assays. Unique analyses like differential protein expression, treatment/control, disease/normal, and post translational modifications can also be carried out with the output of the Microscoop.
| Dimensions (L x W x H) | Electrical controller: 44 cm x 22 cm x 47 cm Optical engine: 68 cm x 46 cm x 22 cm |
| Pattern Segmentation Options | Toolbox for traditional image processing Trained model using Al deep leaming |
| Imagery Wavelength | Dyes: e.g. DAPI, FITC, Cy3, Сy5 Fluorescent proteins: e.g. EBFP2, EGFP, DsRed/mCherry |
| Objectives | 10x (up to NA 0.45) 20x (up to NA 0.80) 40x (up to NA 0.95) |
Orbitrap Fusion Lumos
Orbitrap Eclipse
Orbitrap Exploris 480
Orbitrap Exploris 240
Orbitrap Ascend
Orbitrap Astral
timsTOF Ultra 2
timsTOF Ultra
timsTOF SCP
timsTOF HT
timsTOF Pro 2
ZenoTOF 7600