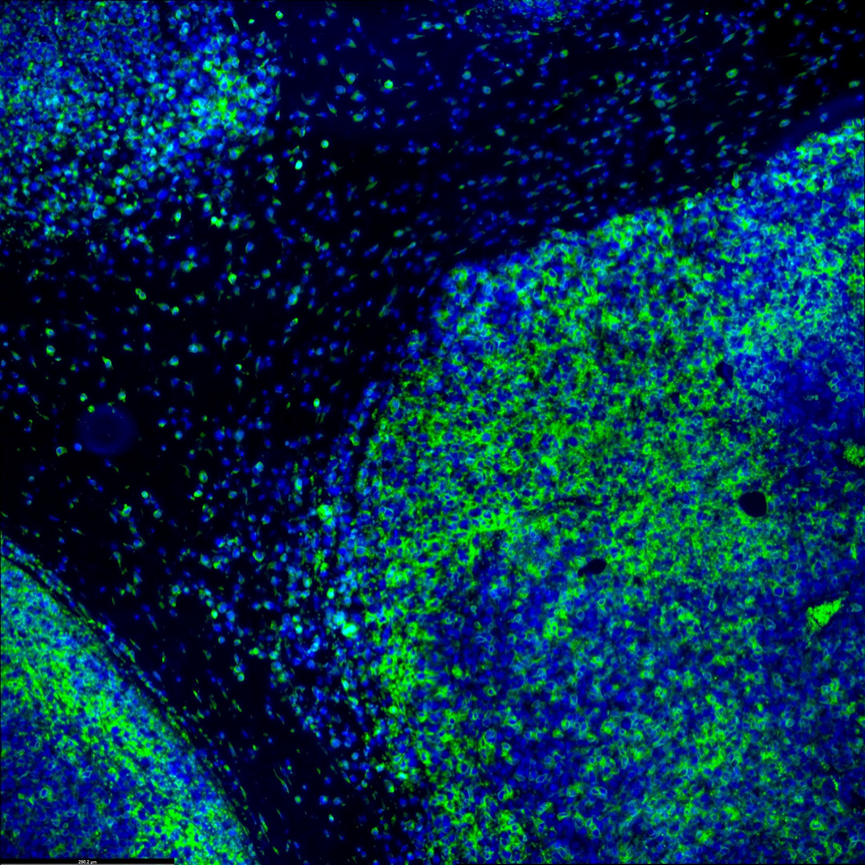
Both inter-tumor and intra-tumor somatic mutations reveal the cancer heterogeneity and can provide crucial information for understanding disease pathogenesis and making clinical decisions. Microscoop™ can further characterize intra- and inter-tumor heterogeneity in addition to information from bulk sequencing or single cell sequencing data to give contextual spatial proteomics atlas to further assess how different phenotypes are associated with clinical variables.
Immune response and tumor microenvironment could greatly affect immunotherapy effectiveness. Specific knowledge of the spatial relationship between immune cells and tumor cells will be important for finding prognostic biomarkers and give a better understanding of the mechanism of treatment resistance. Microscoop™ can be used to study different immune cell subtypes in tumor microenvironment and their cellular neighborhood to associate myeloid cells, lymphoid cells and tumor cells changes with immunotherapy treatment benefits. This new tool can help discover novel biomarkers by tissue compartments or cell types to better understand primary resistance or response to immunotherapies.
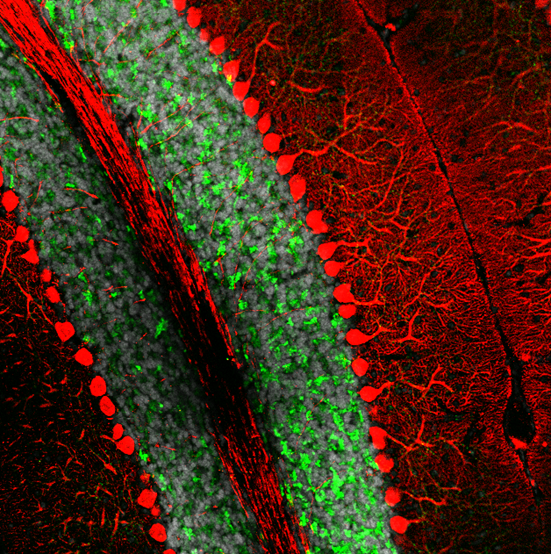
Spatial proteomic studies can help by revealing detailed protein distribution in a complex brain structure. For example, it is well known that damage, degeneration or loss of neurons in cerebellum, results in ataxia. Microscoop™ can be used to profile Purkinje cells proteome to identify changes that occur in these neurons at the time of ataxia appearance. The goal is to identify cellular mechanisms in the cerebellum that specifically target the degeneration of Purkinje cells and find new therapeutic targets.
The image depicts the Purkinje neuron (in red) and granule cell layer (in green) in a mouse cerebellum.
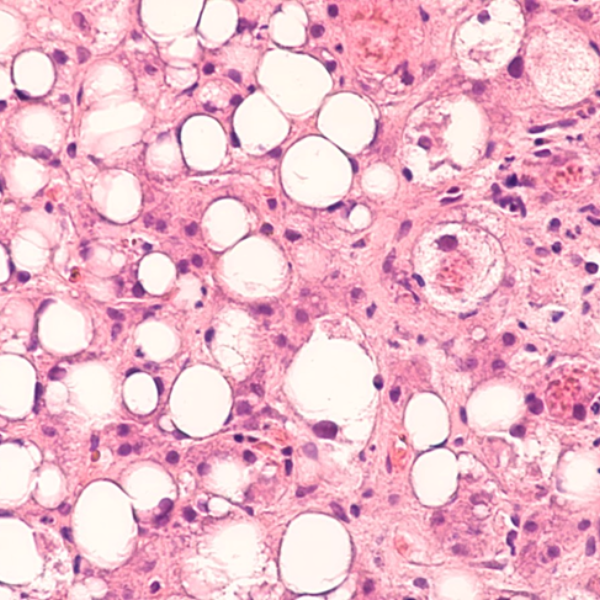
Immune-mediated inflammatory diseases comprise a large number of chronic inflammatory conditions. Optimized anti-inflammatory therapy is therefore essential in the management of these disorders, mainly driven by a dysregulated immune response that initiates and perpetuates the inflammatory reaction. For example, Microscoop™ can be used to study spatial proteomics in both healthy and steatotic livers to map the spatially resolved cellular niches and crosstalks within the microenvironments of hepatic cells, Kupffer cells and lipid-associated macrophages to better understand Nonalcoholic Steatohepatitis(NASH) progression and identify new therapeutic targets.
Microbial pathogens have evolved numerous mechanisms to hijack host’s systems causing disease and lingering side effects. Applying spatial proteomics technology can help build a molecular map of pathogen infection and disease progression by defining the cellular mechanisms of pathogen entry into hosts cells, their replication and transmission, as well as the host defense mechanisms against pathogens. For example, Microscoop™ can be used to study the binding of the SARS-CoV-2 spike protein to its receptor, angiotensin-converting enzyme 2 (ACE2), and subsequent membrane fusion to provide the structural and cellular foundations to understand the multistep entry process and identify other potential therapeutics targeting SARS-CoV-2 entry mechanisms.
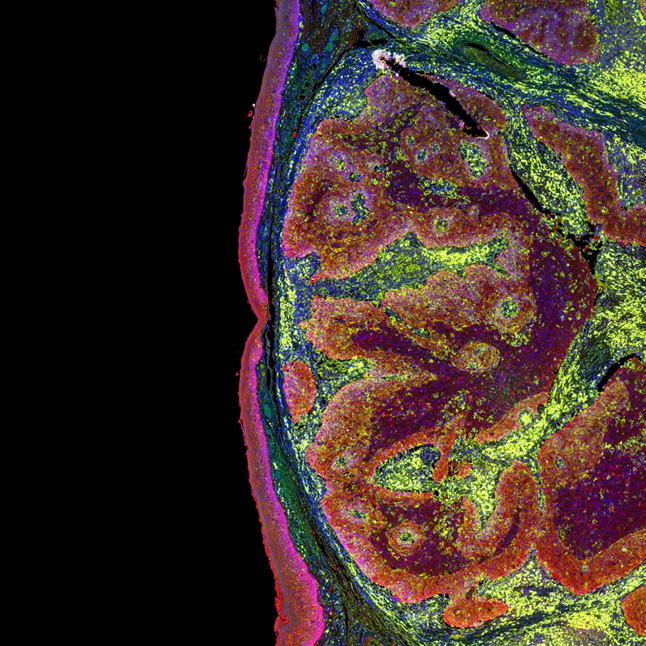
Study the spatial distribution and measure the relative abundance of hundreds of protein analytes from cell types across different developmental stages is critical to the understanding of the dynamic embryonic development processes. Microscoop™ can be used to locate a specific cell layer and track proteome changes in different stages over time to get functional insights into an organism development.