FAQ
General Inquiries
Microscoop™ is a novel spatial biology platform offers unique capabilities for spatial proteomics studies. The technology platform can be applied to both cell- and tissue-based samples, making it broadly applicable to many basic and clinical research fields including oncology, immunotherapy, neuroscience, immunology, infectious disease, and developmental biology.
Microscoop™ Instrument
SYNCELL will offer Microscoop™ to limited customers in the US and Taiwan through an early access program beginning in Q1 2022. The full commercial release is scheduled for early 2023.
The Microscoop™ platform includes an electrical control module, an optical module, a two-photon microscope and a computer.
Image Processing
For very small subcellular structures, it might be necessary to develop a custom AI image processing algorithm for best photolabeling results. However, many of the cellular and subcellular structures can be efficiently recognized by traditional computer visioning tools included in our image processing software toolbox.
Many of the cellular and subcellular structures can be efficiently recognized by traditional computer visioning tools. Please read our user manual to find suggestions on how to adapt your imaging program to the Microscoop™ or call our customer support for more information.
The images one uses to develop an AI algorithm should be representative of the actual cell or tissue samples that will be used for photolabeling. Avoid using confluent cell culture slides or cut tissue sections with very uneven thickness or folds.
Training with more images will improve the accuracy of an AI algorithm to identify desired structures and patterns. We have used as few as 10 images to several hundred images to successfully train a custom AI algorithm.
Microscoop™ user interface (UI) software includes a computer visioning toolkit for users to choose imaging modules that are suitable for their projects. We also offer an image processing training webinar to help users jump start their imaging program. In addition, we offer advanced training classes (for one person or a group) as well as customized services (as part of the GRASP customer service program) to develop imaging solutions tailored to user’s project.
Sample Selection and Workflow
We typically grow our cell cultures on chamber slides (with glass bottom, thickness <170 μm). Tissue sections can be collected on 2-6 slides.
Fixed cells and FFPE (formalin-fixed, paraffin-embedded) or frozen tissue sections are all suitable for use with the Microscoop™.
For FFPE tissues, we typically use 5-10µm sections.
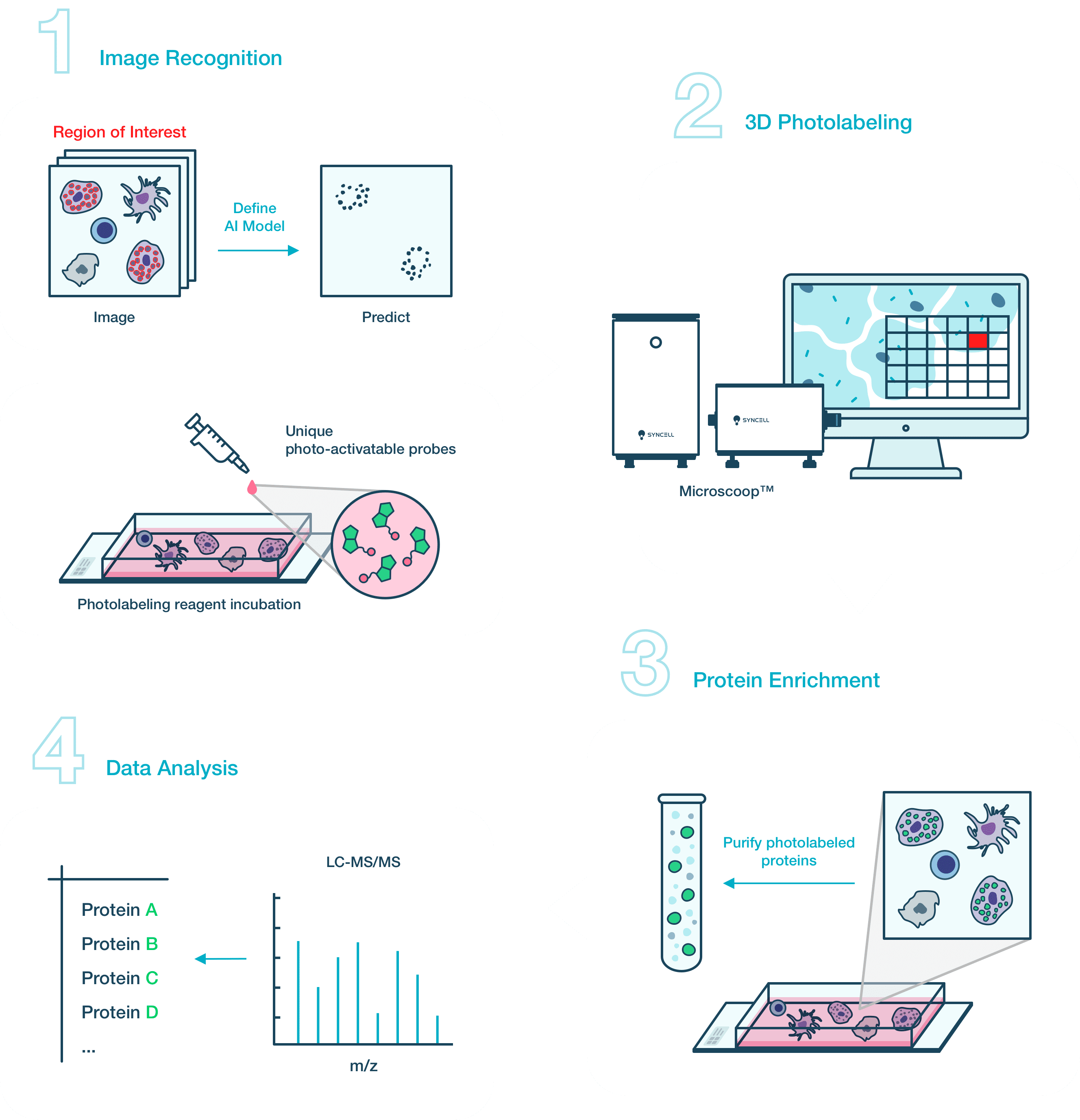
The photolabeling time is application-driven. If the targeted structure is greater than 3 μm in both length and width, and at least one targeted structure is present in a cell, it will take about 24 hours to label 4×105 cells to collect enough cells for a LC-MS analysis. If the targeted structure is finite and sparse in the targeted sample area, it will take more sample slides and longer photolabeling time. Typically, we suggest starting with 1×106 cells to optimize the workflow from photolabeling to LC-MS analysis for this type of sample.
Nanoscale liquid chromatography system coupled with a middle or high-end mass spectrometer (MS) is recommended for the analysis of Microscoop™ photolabeled samples. If the targeted structure is greater than 3 μm in both length and width, and at least one such structure is present in a cell, the samples can be analyzed using a MS with ~10 Hz of MS/MS acquisition rate, such as Q-exactive™ Plus (Thermo Fisher Scientific). When the targeted structure is tiny or sparse, we recommend analyzing the sample with a MS which features >20 Hz of MS/MS acquisition rate, such as Orbitrap Fusion™ Lumos™ (Thermo Fisher Scientific), Orbitrap Eclipse™ (Thermo Fisher Scientific), timsTOF (Bruker), TripleTOF® (SCIEX), ZenTOF (SCIEX).